Normalization using VST
STEP9 Data processing
The first step in data processing is the process of normalization, in this case we are performing a log transformation of the data and scaling it to a factor of 10,000
# Normalize and identify variable features for each dataset independently
pbmc <- NormalizeData(pbmc, normalization.method = "LogNormalize", scale.factor = 10000)
#OUTPUT
Performing log-normalization
0% 10 20 30 40 50 60 70 80 90 100%
[----|----|----|----|----|----|----|----|----|----|
**************************************************|
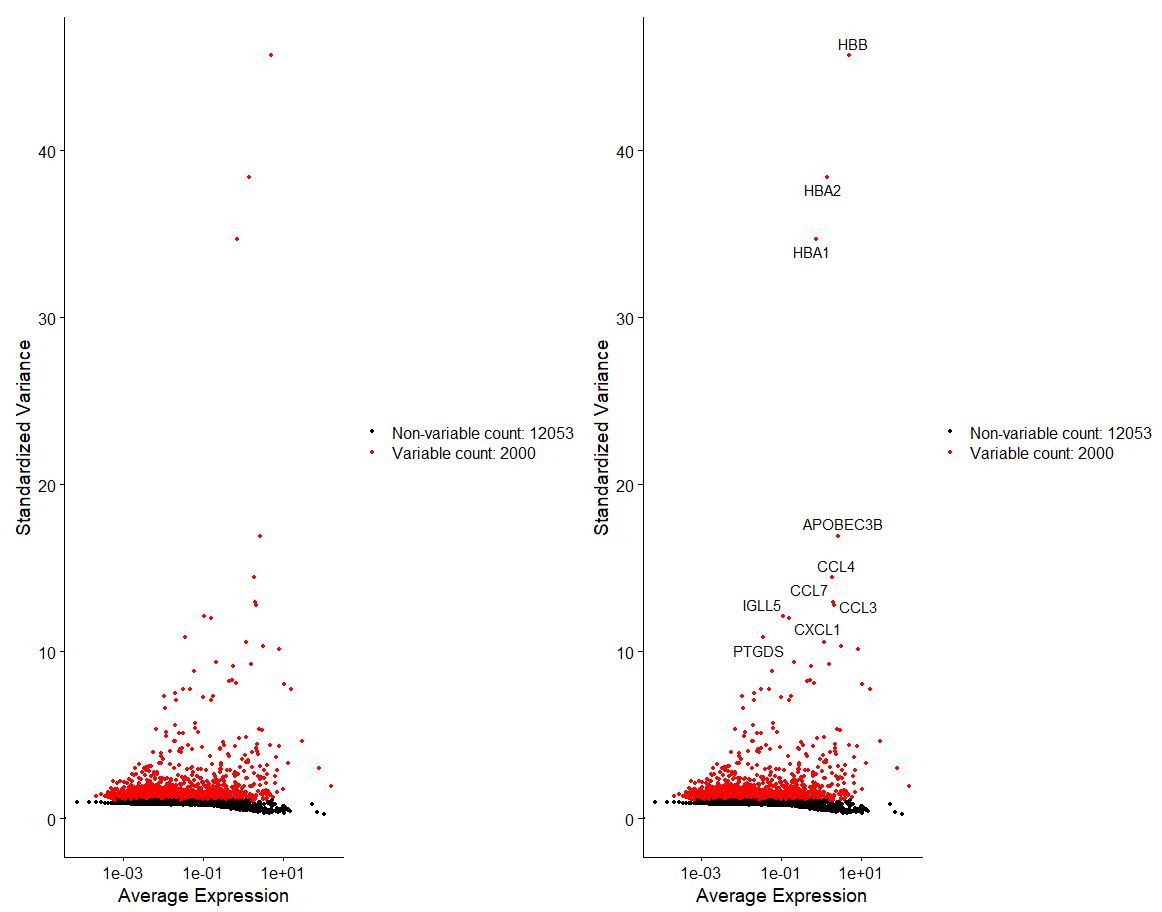
Now we want to select for most variable features in our dataset, or in this instance the top 2000 most variable features.
# select highly variable features
pbmc <- FindVariableFeatures(pbmc, selection.method = "vst", nfeatures = 2000)
#OUTPUT
Calculating gene variances
0% 10 20 30 40 50 60 70 80 90 100%
[----|----|----|----|----|----|----|----|----|----|
**************************************************|
Calculating feature variances of standardized and clipped values
0% 10 20 30 40 50 60 70 80 90 100%
[----|----|----|----|----|----|----|----|----|----|
**************************************************|
# Identify the 10 most highly variable genes
top10 <- head(VariableFeatures(pbmc), 10)
top10
#OUTPUT
[1] "HBB" "HBA2" "HBA1" "APOBEC3B" "CCL4" "CCL7" "CCL3" "IGLL5" "CXCL1" "PTGDS"
# plot variable features with and without labels
plot1 <- VariableFeaturePlot(pbmc, assay = "RNA")
plot2 <- LabelPoints(plot = plot1, points = top10, repel = TRUE, xnudge = 0, ynudge = 0)
plot1 + plot2
#OUTPUT
Warning messages:
1: Transformation introduced infinite values in continuous x-axis
2: Transformation introduced infinite values in continuous x-axis
Next, we are going to perform an action that scales and centers features in the dataset.
# Scale data
all.genes <- rownames(pbmc)
pbmc <- ScaleData(pbmc, features = all.genes, verbose = TRUE)
#OUTPUT
Centering and scaling data matrix
|===========================================================================================================| 100%