Experimental Setup
Perhaps one of the most central aspects of scRNAseq studies in the study design. In the opinion of this instructor, the study design is the most fundamental aspect of scRNAseq experiments. One of the reasons for stringent study design are technical variability. Technical variability can give rise to varying cell capture efficiencies, varying library quality, batch effects due to underlying biology or even experimental handling and amplification bias due to chemistry or transcript. This highlights the problem of confounding. When an external factor distorts the effect and leads to a mixing of effects between the treatment and control we term that phenomena as confounding. Confounding can predominnatly be attributed to batch effects and inappropriately designed experiments. This is highlighted in the figure below:
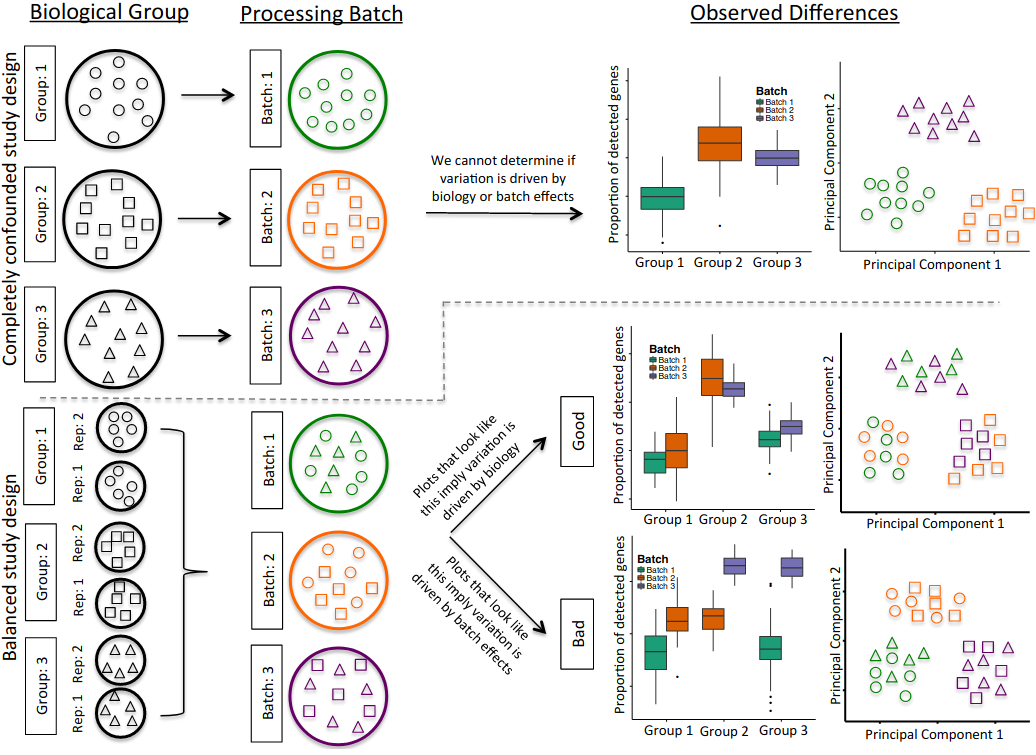 Figure adapted from Hicks et al., 2018: Biostatistics
Figure adapted from Hicks et al., 2018: Biostatistics
It is absolutely essential, to never compromise on your “n”. Have atleast n=3 for each of your batches (bare minimum) however n>3 is advisable. If you are trying to look at population wide effects then performing statistics on single cells is not advised and a process of pseudobulking is recommended where all reads are compressed on the basis of cell type. So in that case a n=1 or n=2 will be completely inappropriate from the context of a study design. An appropriate mixed model study allows for multiple sample to be sequenced where biological variability and technical variability is addressed at multiple layers of batch correction.