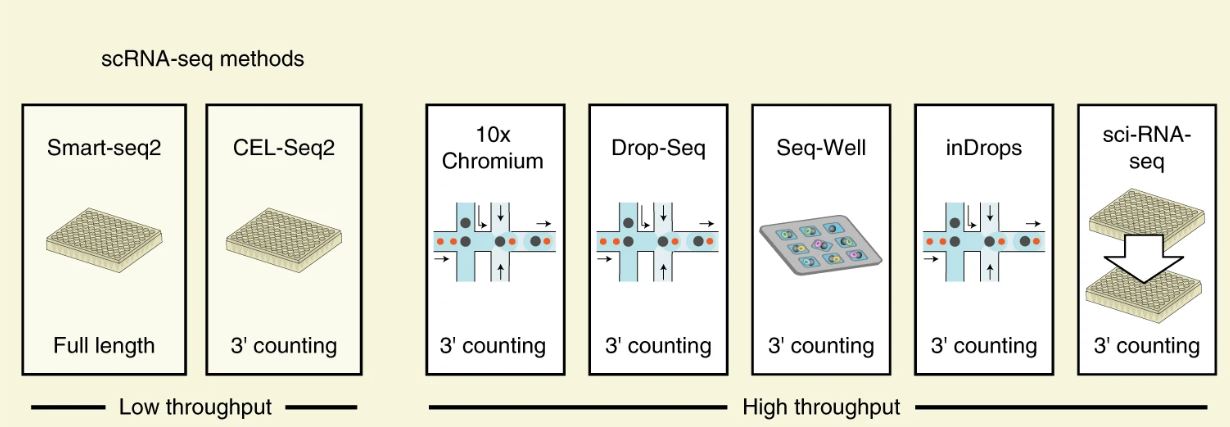
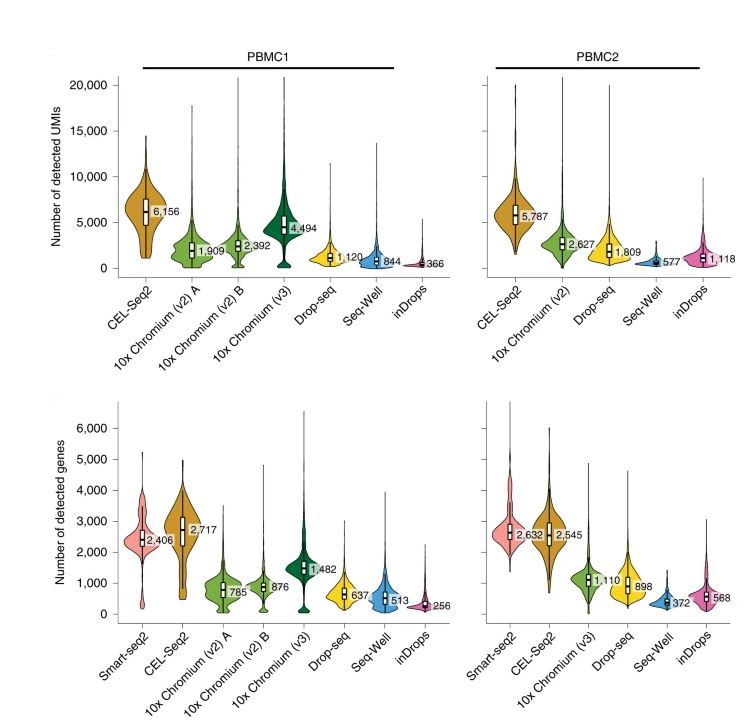
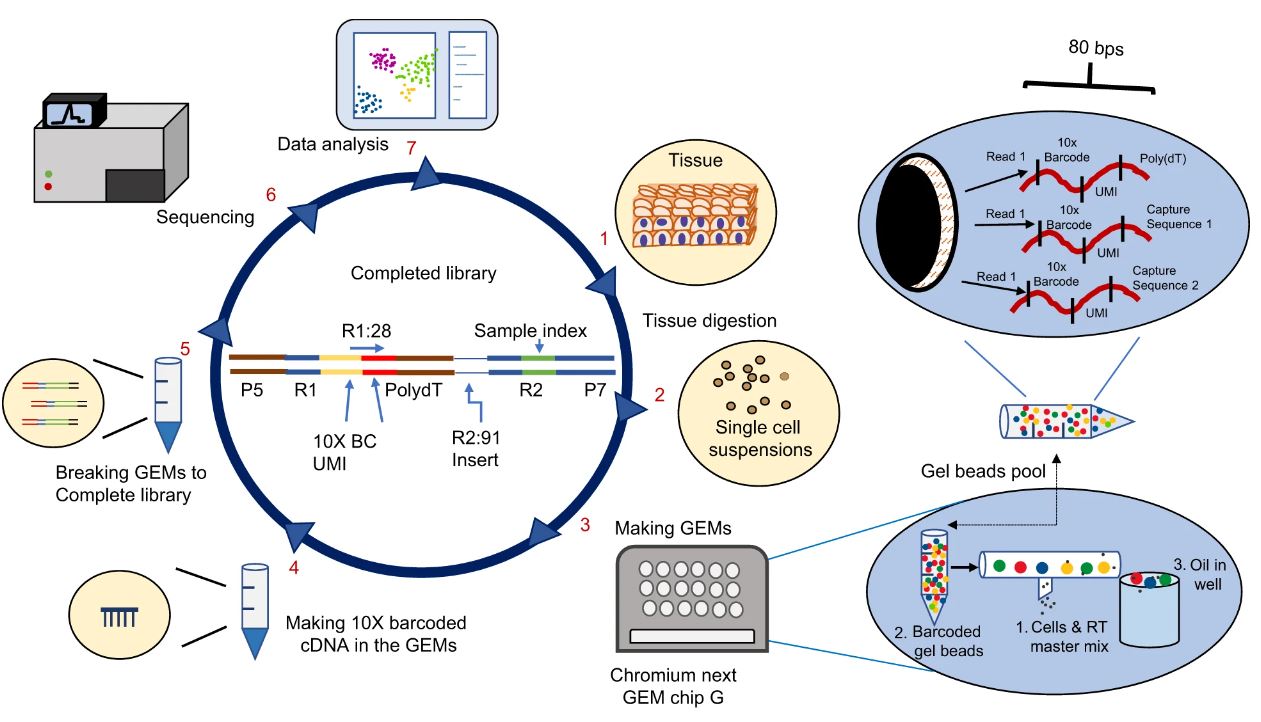
Library Preperation
Library preperation involves the creation of a partitioned single cell library which allows for the sequencing of RNA transcripts (cDNA) in tandem across multiple single cells. The modality for library prep is an important consideration as it drives overall sequencing quality. Nevertheless most appropriate library/sequencing platform depends on the biological question. For example, if one is interested in characterizing the composition of a heterogeneous tissue, then a droplet-based method is more appropriate, as it allows a very large number of cells to be captured in a mostly unbiased manner. On the other hand, if one is interested in characterizing a specific cell-population for which there is a known surface marker, then it is probably best to enrich using FACS and then sequence a smaller number of cells at higher sequencing depth.
Figure adapted from Ding et al., 2020: Nature Biotechnology
Still, across multiple platforms differences exsit with full-length seqnecing being the most appropriate especially with regards to identifying transcript isoforms. However if you are intrested in evaluating or discovering cells of a small number and rarity, then using a UMI based approach is appripriate as it allows you to sequence multiple cells. It is always advised to go over studies on multiple sequencing technologies to judge which method suits your question. Here at Tulane we use the 10X platform with the V3 chemistry, which is optimal with regards to UMI and cells-sequenced yields.
Figure adapted from Ding et al., 2020: Nature Biotechnology
As of 6/15/2023 the Tulane sequencing core utilizes the 10X Genomics platform using V3 chemsity. The overall workflow of the 10X stsem involves the generation of single cell partitioned libraries, which are then pooled and sequenced in tandem in a HiSeq2500 sequencer. Although staff at the sequencing core generate libraries for you, it is not a bad idea to go over the 10X manuals to study how single cell libraries are constructed, so you can have a better understanding of how cell quality (>90% viability, preferably 95% viability) is essential for high quality data.
Figure adapted from Li et al., 2021: IJOS